Microhaplotypes
We identified haplotypes in kelp rockfish that occur over a very small genetic distance (~130 bp). The key to using such a small genomic region is that we can quickly and easily genotype thousands of fish using amplicon sequencing.
We have tested our rockfish microhaplotypes across 49 Sebastes species from the NE Pacific, and are able to distinguish species identities using these genetic markers.
Although other studies are beginning to use haplotypes for population structure and ancestry, using microhaplotypes for pedigree questions – parentage and sibship analysis – provides a much greater increase in power over single SNPs.
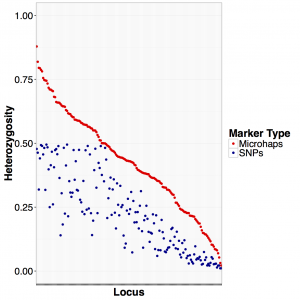
Using heterozygosity as a measure of statistical power, this figure shows the heterozygosity of the microhap comprised of all SNPs in a locus compared to the single SNP with the highest minor allele frequency in that same locus. Bi-allelic SNPs have a maximum heterozygosity of 0.5 across all 171 loci tested. We settled on a set of 96 loci for genotyping rockfish in this project.